Newsroom

Learn more about Vela Diagnostics through the latest press releases, events and more.
If you are a member of the media with an enquiry about Vela Diagnostics and related products, feel free to get in touch.
More than 20 companies in Singapore given green light to make COVID-19 test kits | Video


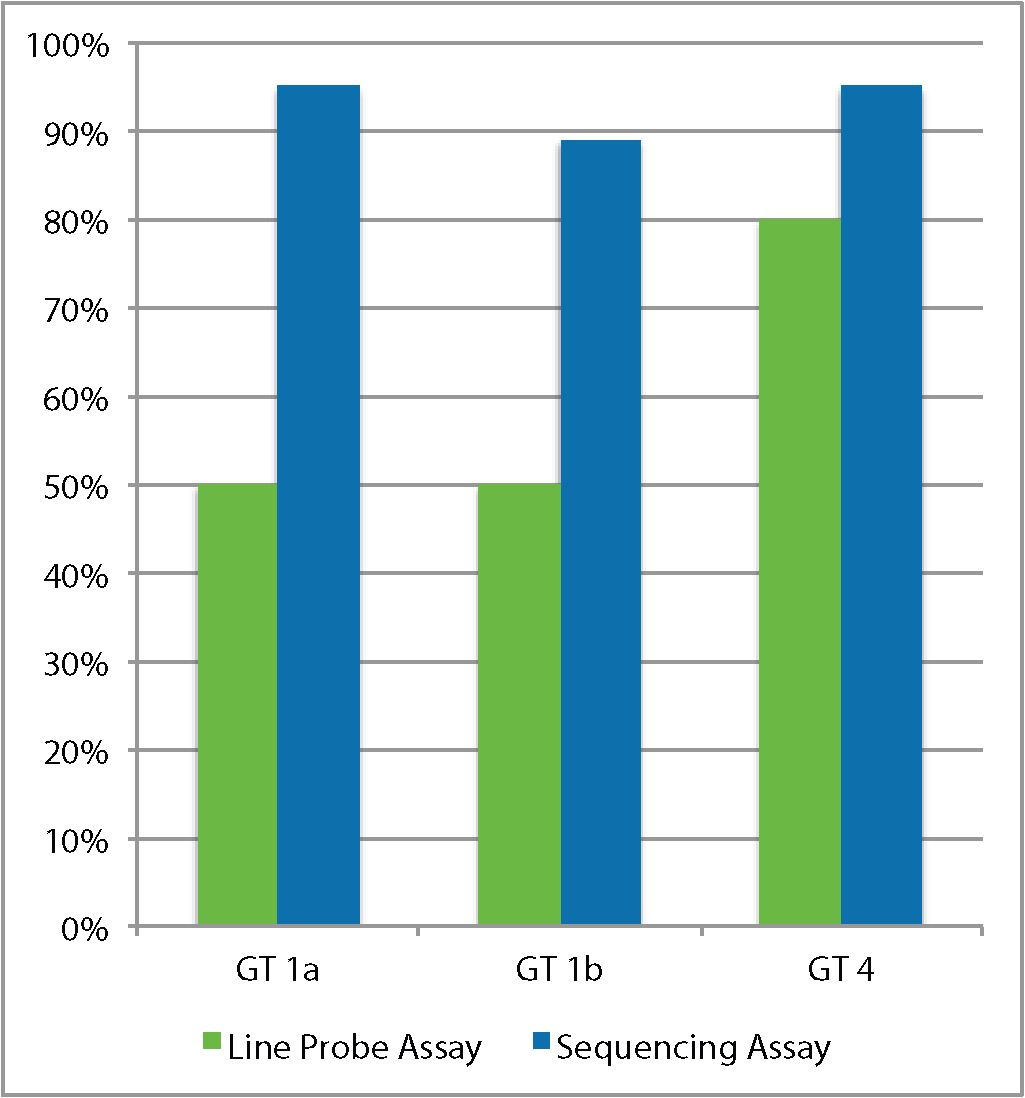
The Sentosa SQ HCV Genotyping Assay is a clinically validated Next-Generation Sequencing (NGS) test for genotyping and variant calling for Hepatitis C Virus (HCV) in clinical samples*.
- Achieves 100% genotyping correctness
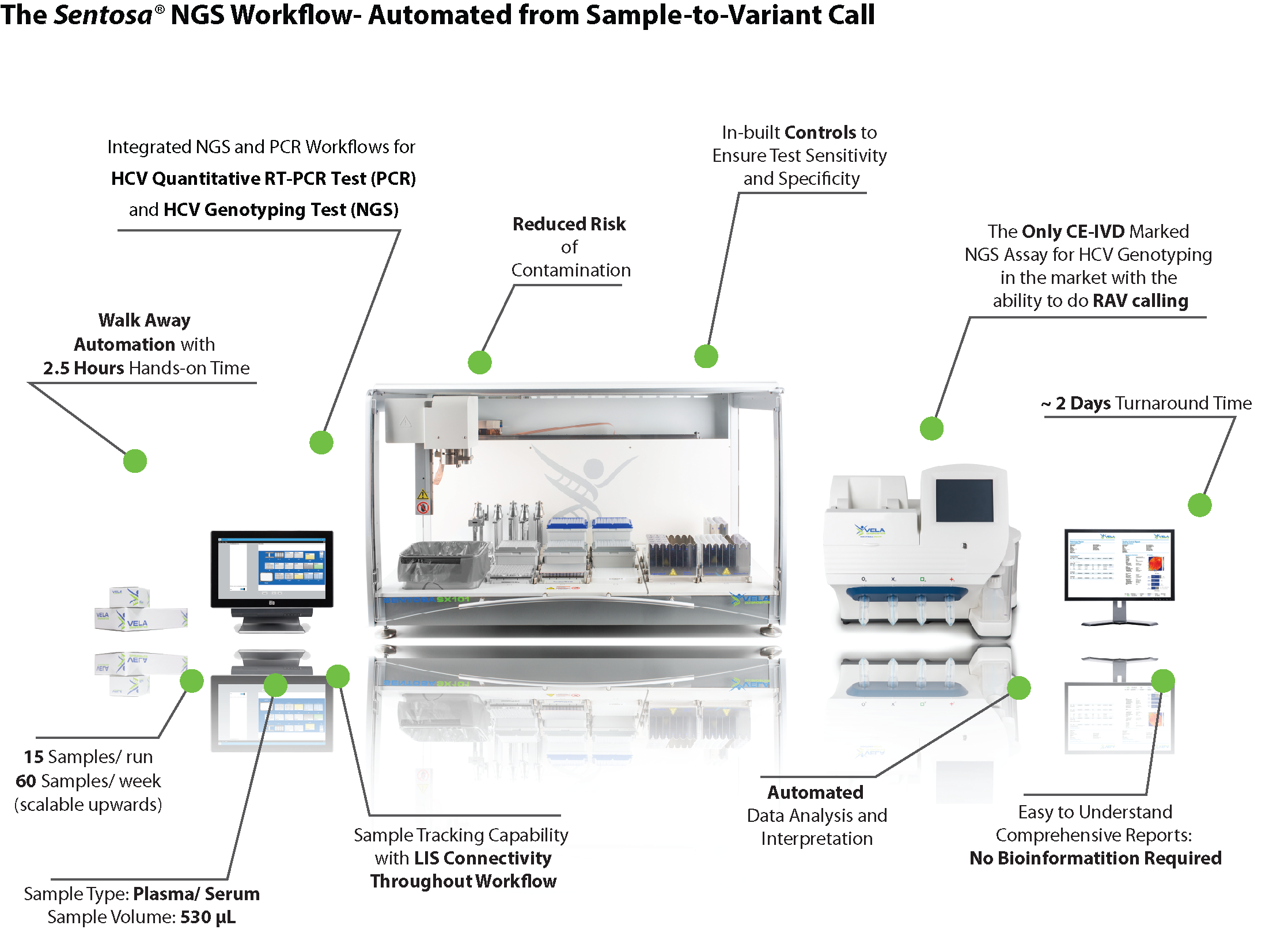
- Enables highly reproducible results for routine diagnostics, through automation of sample processing, library prep, sequencing, result reporting on a single platform
- Validated and approved for in vitro diagnostic use (CE-IVD)*
 |
 |
 |
| How does deep sequencing increase treatment efficacy as compared to line probe genotyping? | Solve more cases, in a shorter time with our automated end-to-end NGS solution | Request for the full list of HCV Resistance Associated Variants (RAVs) which are in line with the European and American Liver Guidelines |
| Read More |
Read More |
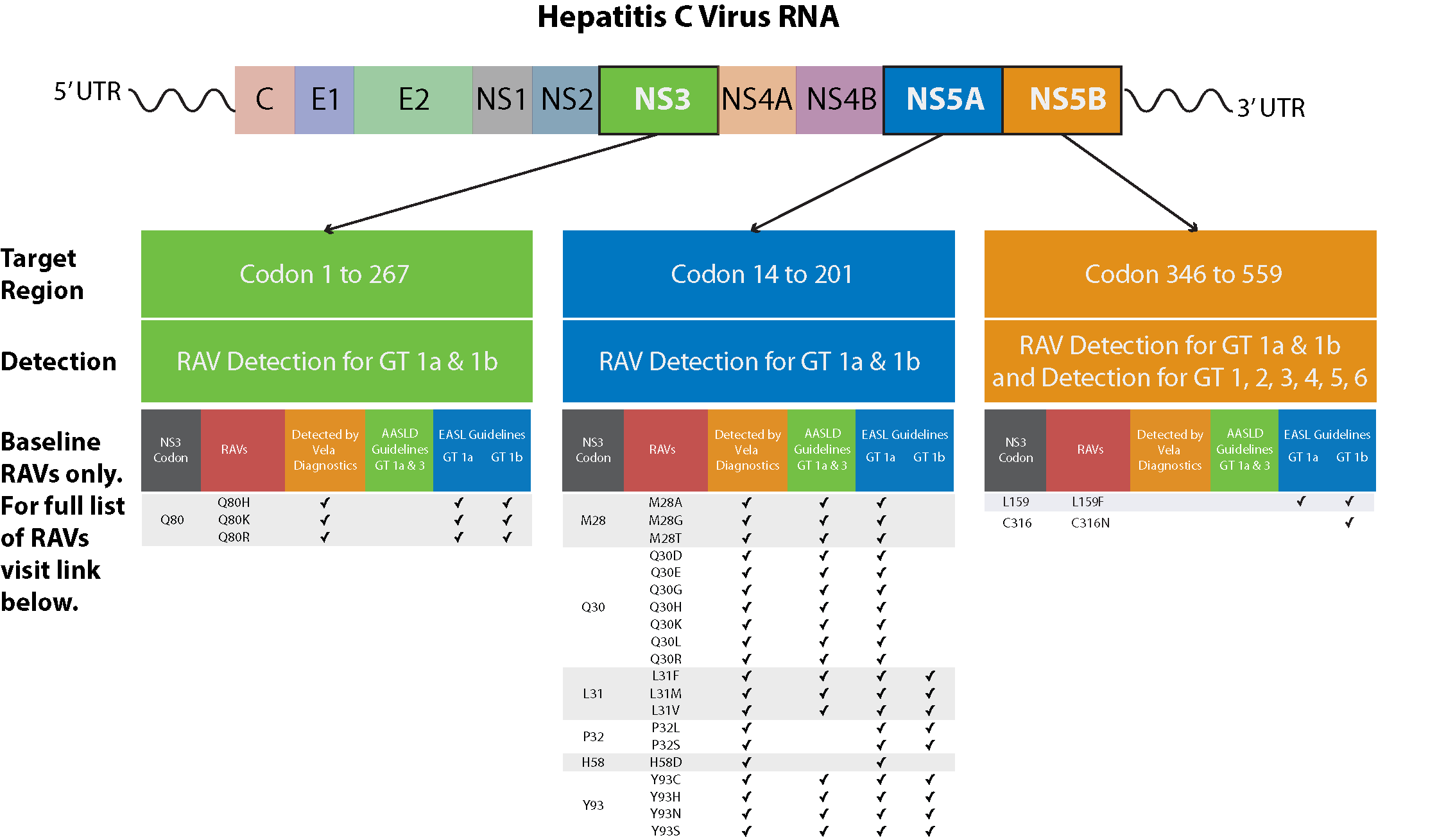
Request |
 |
 |
|
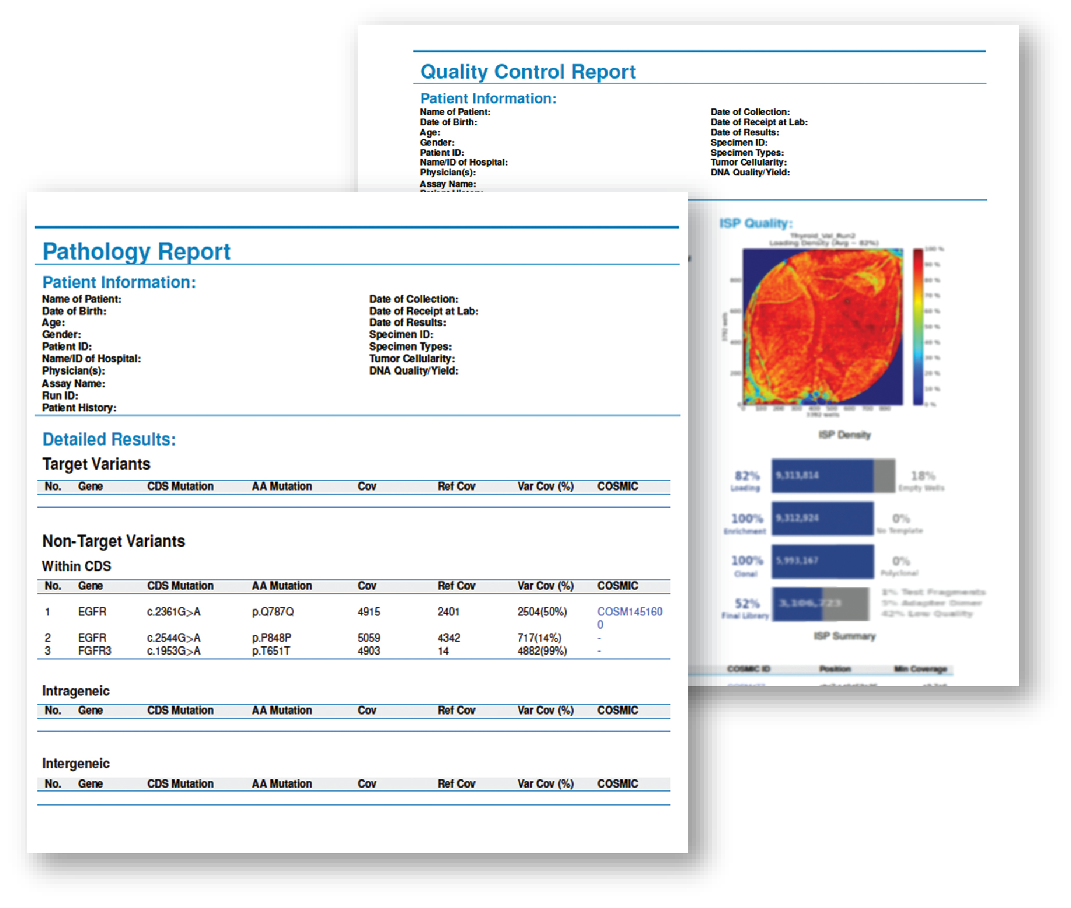
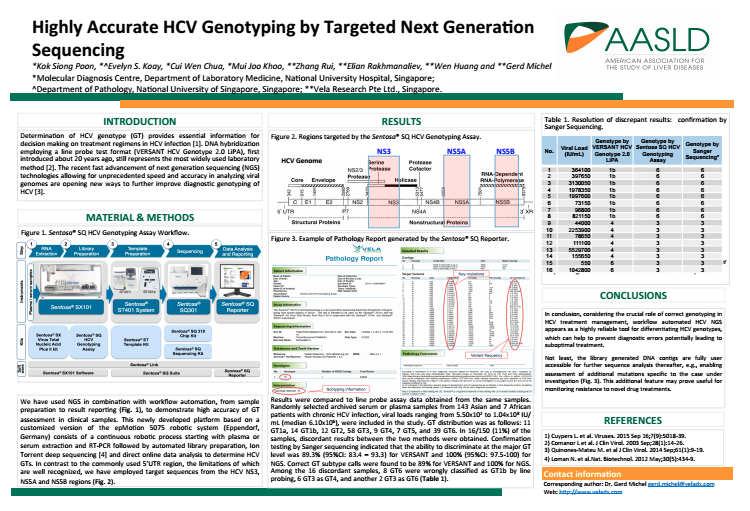
| Request a sample report-comprehensive reports automatically generated by the Sentosa SQ Reporter. Simple to read, easy to understand. | Download a scientific poster- Highly accurate HCV genotyping by targeted Next-Generation Sequencing. | |
| Request |
Download |
*only RUO version is available in the US

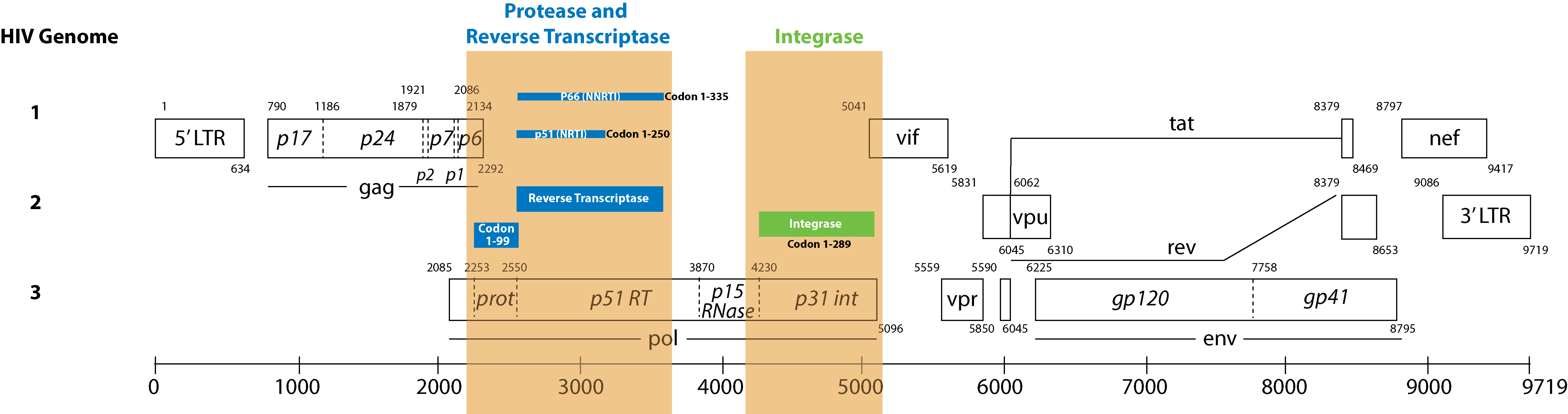
The Sentosa SQ HIV Genotyping Assay detects drug resistant mutations in HIV positive samples from plasma, to rapidly detect mutations in the relevant regions of Protease, Reverse Transcriptase and Integrase genes.*
- Overall mutation detection correctness: 99.06%
- Assay reproducibility: 100%
- Interpretation report generated by Sentosa SQ Reporter is based on Stanford HIV database
-
Targeted mutation variant frequency of 20% can be detected by assay at 1000 copies/mL;Targeted mutation variant frequency of 5% can be detected by assay at 4000 copies/mL
-
Codon coverage:
- Protease: 1 - 99
- Reverse Transcriptase: 1 - 376
- Integrase: 1 - 288
 |
 |
 |
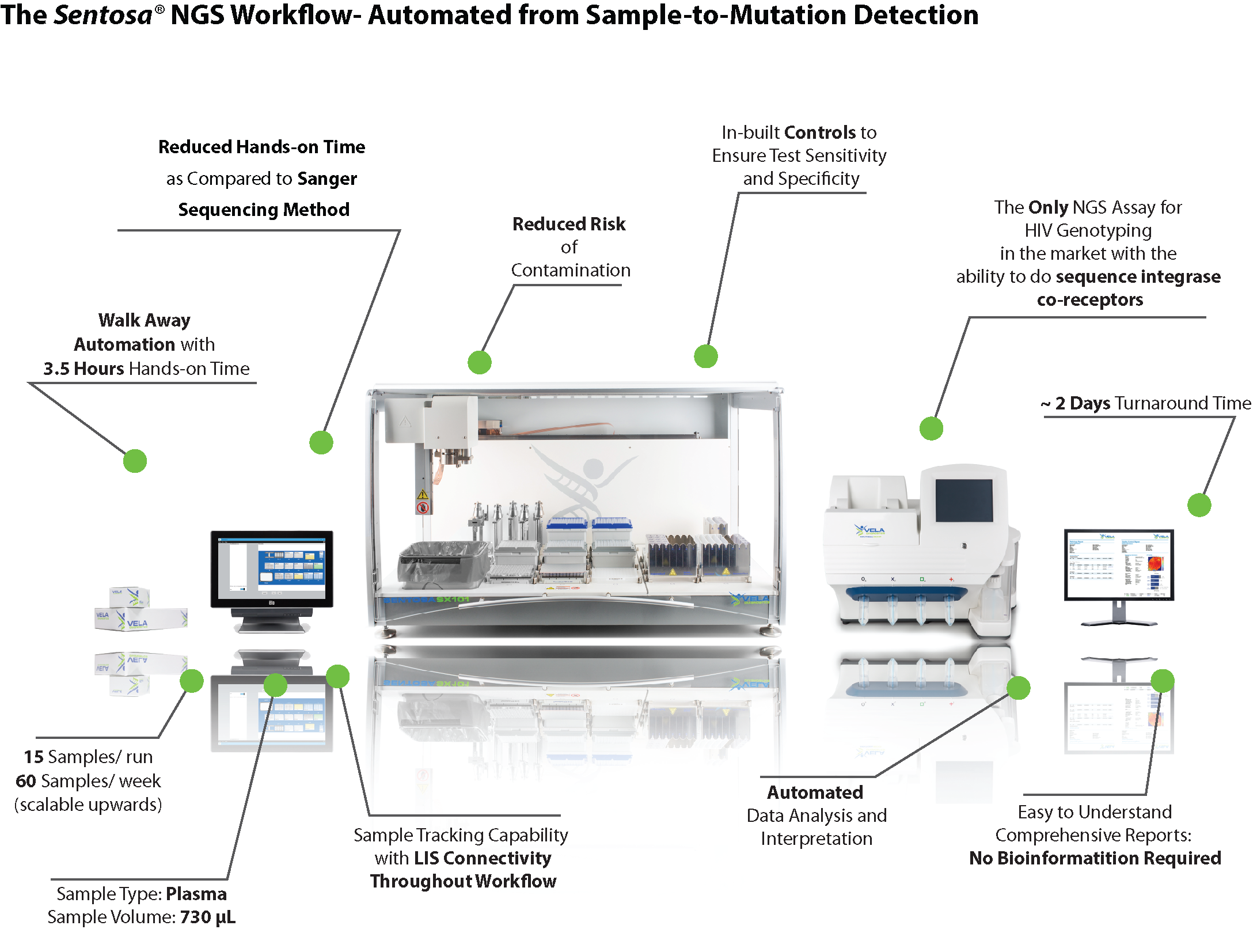
| The HIV Genome- The Sentosa SQ HIV Genotyping Assay provides simultaneous detection of major drug resistant mutations, targeting first line HIV treatment*. | Solve more cases, in a shorter time with our automated end-to-end NGS solution | Request a sample report-comprehensive reports automatically generated by the Sentosa SQ Reporter. Simple to read, easy to understand. |
| Read More |
Read More |
Request |
 |
||
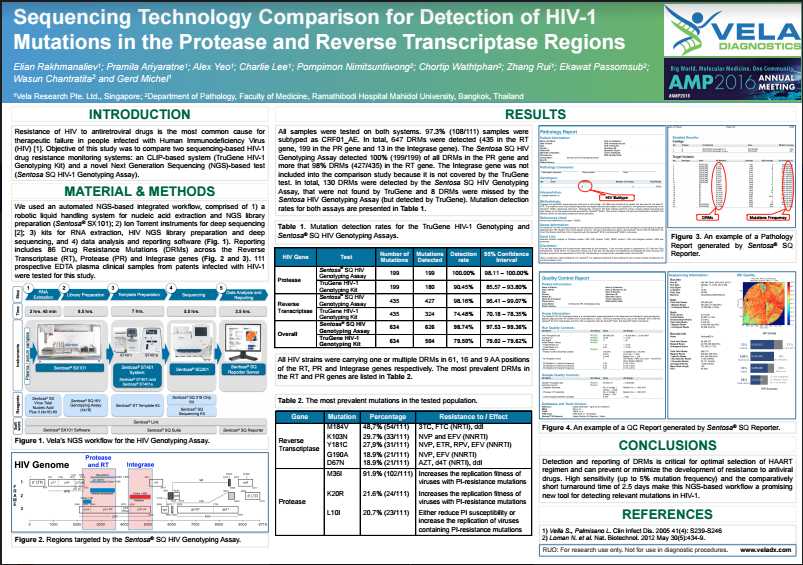
| Download a scientific poster-Detection of HIV-1 mutations in the protease and reverse transcriptase regions. | ||
| Download |
*only RUO version is available in the US
|
Live Webinar on Genomeweb This online seminar will highlight recent advances in the use of next-generation sequencing to detect drug-resistant mutations in patients with HIV or HCV.* Sanger sequencing and PCR have been the standard molecular methods in clinical diagnostics for decades, but NGS is now swiftly becoming a routine method in different areas of clinical diagnostics, including treatment management for viral diseases. Title: An NGS Workflow for HCV and HIV Resistance Testing Date: March 2, 2017 (Thursday) Time: 1:00 pm USA (Eastern Standard Time GMT-05:00) *Speakers used a CE-IVD product, only RUO version is available in the US |
|
|
On-Demand Seminar 4 distinguished speakers discuss HCV and HIV genotyping using Next-Generation Sequencing at the 19th Annual Meeting of the European Society for Clinical Virology (ESCV) Lunch Symposium*:
*Speakers used a CE-IVD product, only RUO version is available in the US |
|






